Relationship between the presence of mycobacteria and non-mycobacteria in metalworking fluids
Drs. Frederick J. Passman, Katalin Rossmoore & Leonard Rossmoore | TLT Case Study March 2009
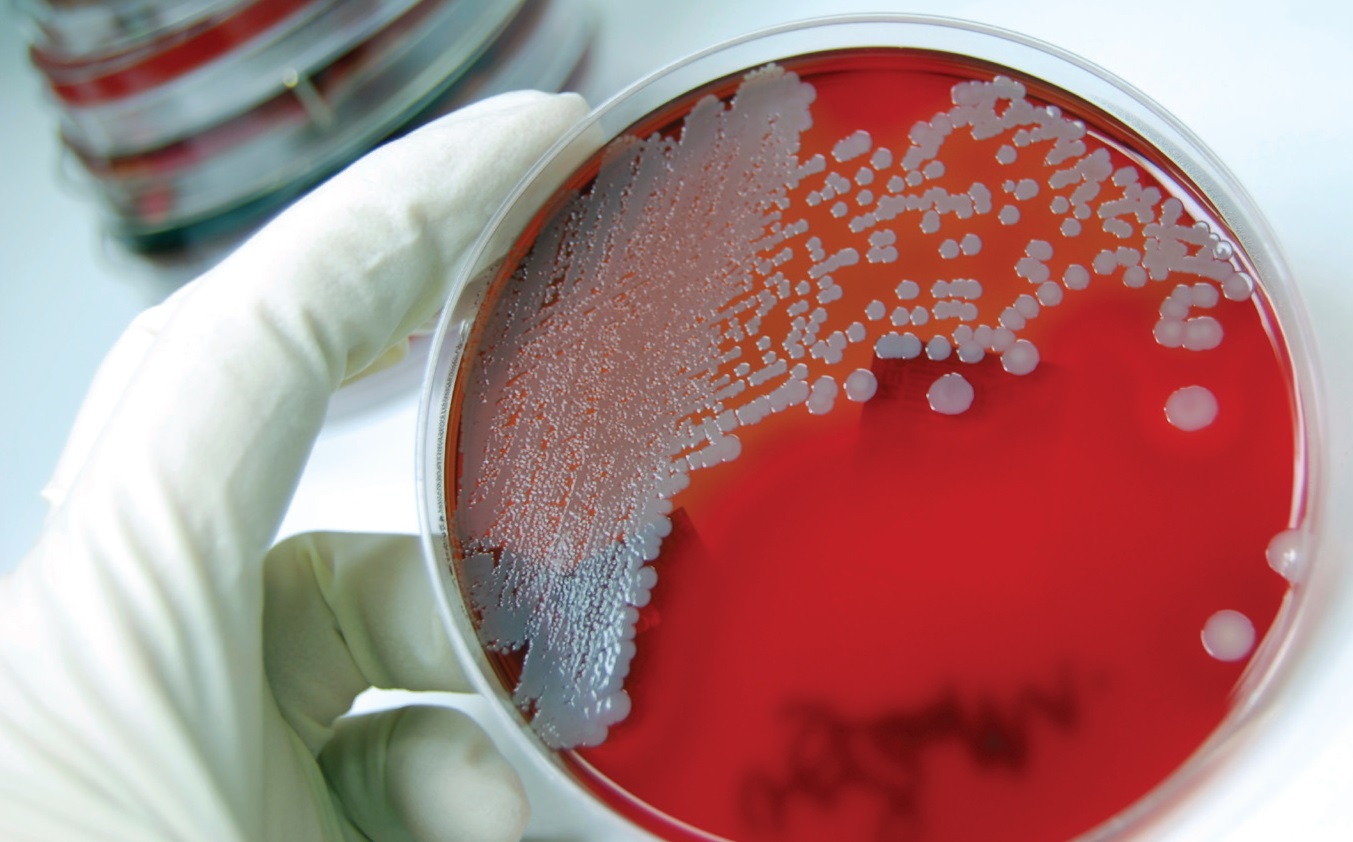
Researchers investigate test samples to get a better understanding of the microbial population in MWF.
KEY CONCEPTS
• Fewer than 200 cases of hypersensitivity pneumonitis (HP) have been reported among workers that have been exposed to MWFs.
• Colonies aren’t visible to the naked eye until they contain approximately two billion cells.
• If future HP outbreaks occur, a multidisciplinary approach is recommended to the cause and effect investigation.
This case study reports culture and acid-fast bacteria direct counts from 99 MWF samples received for microbiological testing at Warren, Mich.-based Biosan Laboratories between December 2006 and September 2007. The objective of the study was to determine whether the presence of gram-negative bacteria affected the presence of mycobacteria in MWF. Data from the MWF tested during this investigation demonstrated that there is a statistically significant relationship between mycobacteria recovery and non-mycobacteria recovery in MWF.
BACKGROUND
Mycobacterium immunogenum is a relatively fast-growing acid-fast bacterium that has been recovered from some MWF systems nearby where workers contracted the respiratory disease hypersensitivity pneumonitis (HP). Since the sentinel HP outbreak at a metalworking plant in 1991, there have been fewer than a dozen HP clusters reported in metalworking facilities (1). In total, fewer than 200 cases of HP have been reported among workers exposed to MWF. These statistics don’t diminish the seriousness of the HP issue. Rather, they highlight the difficulty of assessing cause-and-effect relationships. Further complicating diagnostic efforts is the fact that most of the HP clusters included four to 10 individuals at plants where 100-500 workers were exposed to MWF aerosols routinely. In their survey of HP outbreaks, Kreiss and Cox-Ganser noted that mycobacteria had been recovered at five of eight plants where HP had been reported between 1991 and 1996.
Despite the absence of mycobacteria from a significant number of the facilities at which HP clusters were reported and the fact that a number of gram-negative bacteria and fungi commonly recovered from MWF are known to cause HP (2), a number of investigators argued that M. immunogenum was the organism responsible for HP at MWF facilities. The apparent inverse relationship between mycobacteria and non-mycobacteria colony count data in MWF samples was used in support of this theory. Watt argued that the use of certain formaldehyde condensate microbicides selectively inhibited the growth of gram-negative bacteria and thereby created conditions particularly favorable for the proliferation of mycobacteria in MWF (3).
There was one major problem with all of the early data. Before 1998 the medium used to enumerate mycobacteria in MWF did not contain antibiotics to inhibit the growth of non-mycobacteria. Consequently, when non-mycobacteria were present, they would overgrow the culture medium before mycobacteria colonies could be seen. Colonies don’t become visible to the naked eye until they contain approximately two billion cells. Assuming that each colony is formed from a single cell, it takes 30 generations (cell division cycles) to get from one cell to a billion. The time required for a cell to divide is called the generation time. A bacterium with a 0.5h generation time will form a visible colony in 15h. One with a generation time of 6h takes 7.5 days to form a visible colony. Many of the commonly recovered gram-negative bacteria have generation times of 0.5 to 1.5h. Fast-growing mycobacteria (non-tuberculosis mycobacteria–NTM) have generation times of 6 to 10h (other mycobacteria grow substantially slower than NTM). Colonies continue to grow (diameter increases) after they become visible so that colonies that are close to one another eventually merge. Confluent growth occurs when all of the colonies on a growth medium merge to form a uniform lawn of growth over the medium’s surface. This can occur within three to four days when the culture medium has more than 100 colonies on its surface.
To illustrate the effect this phenomenon has on the ability to detect mycobacteria in a MWF also contaminated with non-mycobacteria, we use an analogy. Imagine yourself in a helicopter over Times Square on New Year’s Eve. Now try to count the number of children under the age of five who are standing in the crowd. Any child who is not on an adult’s shoulders is eclipsed by the surrounding adults. They can’t be counted. Similarly, colonies of mycobacteria may be present but are eclipsed by the confluent lawn of fast-growing colonies. Once appropriate antibiotics were incorporated into the mycobacteria enumeration media, mycobacteria were recovered with increased frequency from MWF that also supported substantial populations of non-mycobacteria. The apparent inverse relationship seemed to be an artifact of earlier test methodology. Despite anecdotal reports of the relative frequency of mycobacteria recovery, there were no published reports of the relationship between NTM and non-mycobacteria recoveries in MWF.
To address this information gap, the authors decided to test the next MWF samples received by the laboratory until we had data for approximately 100 samples. All samples would be tested for culturable bacteria (non-mycobacteria), culturable fungi, culturable NTM and other acid-fast bacteria. The latter test is a direct count method.
Assuming that each colony is formed from a single cell, it takes 30 generations (cell division cycles) to get from one cell to a billion.
METHODOLOGY
Approximately 100 MWF samples received at Biosan Laboratories between December 2006 and September 2007 were tested by the standard plate count method (4) for heterotrophic bacteria and fungi. Briefly, serial dilutions of MWF sample were plated onto plate count agar (PCA) for bacterial enumeration and PCA augmented with glucose, Chloramphenicol and Gentamycin for fungal enumeration, and MycoAgar augmented with Chloramphenicol, Gentamycin, Cycloheximide and glycerol for NTM enumeration. After inoculation, heterotrophic bacterial and fungal plates were incubated at 30 C and observed for colonies for up to four days. Mycobacteria plates were incubated at 30 C for up to 14 days. Results were reported as colony-forming units (CFU)/mL MWF.
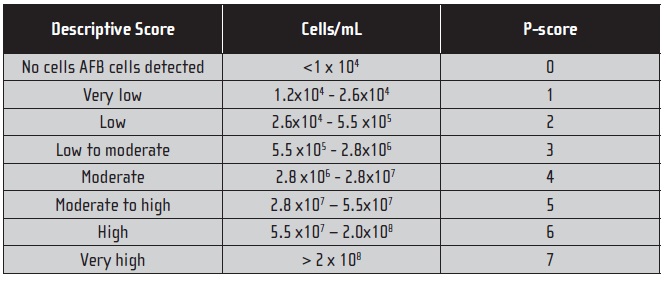
The MWF samples also were examined by ASTM E 2564 (5) to get acid-fast bacteria (AFB) direct counts. Table 1 presents the AFB direct count scoring key.
Table 1. Acid-Fast Direct Count Scoring Key.
Once all of the data were collected, the culture results were transformed to Log10 CFU/mL and all parameters were tested statistically for covariance.
RESULTS
In discussing the test results, we’ll use the term bacteria when referring to non-mycobacteria. Of the 99 samples received during the case study, 79 were positive for at least one of the parameters tested. Bacterial CFU/mL were ≥ 2 in 68 samples and fungal CFU/mL were ≥ 2 in 28 samples. Seventeen of the samples yielded either ≥ 2 CFU NTM/mL, an AFB direct count score of ≥ 1 (<6,000 acid-fast cells/mL, where 6,000 cells/mL is the methods lower detection limit) or both. Seven of the 17 samples (42%) that had detectable numbers of NTM or AFB also had detectable bacteria, and five (32%) had fungi. Approximately 43% of the samples that yielded bacteria also yielded fungi. There was only one MWF sample from which fungi but not bacteria were recovered. All of the test data were compared statistically. Figure 1 presents the NTM and fungal Log CFU/mL, both as functions of the Log CFU bacteria/mL data for all 99 samples.
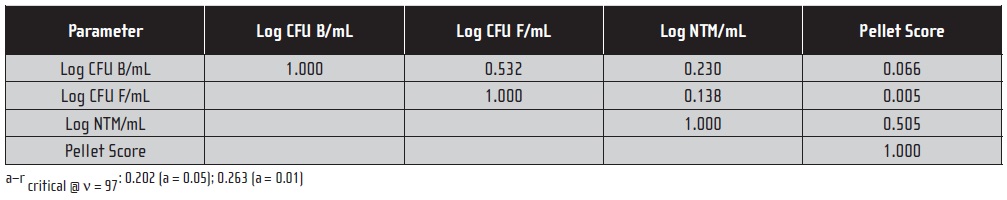
Table 2 shows the correlation matrix. Values of r (correlation coefficient) that are less than the critical values shown in the table’s note are not significant. Bacterial and fungal colony counts covaried significantly, as did NTM colony counts and AFB direct counts. Interestingly, the statistics suggest a slight, positive correlation between bacterial and NTM colony counts (r = 0.230; not significant at the 95% confidence level but significant at the 99% confidence level). It’s most likely that this apparent positive correlation was due to the number of microbial-contamination free samples in the data set. To test this hypothesis, we recomputed the correlation statistics using only data from samples that yielded ≥ 2 CFU NTM/mL. There were 15 samples that met this criterion.
Table 2. Correlation Matrix for Microbiological Parameters from 99-Sample Metalworking Fluid Data Set (a).
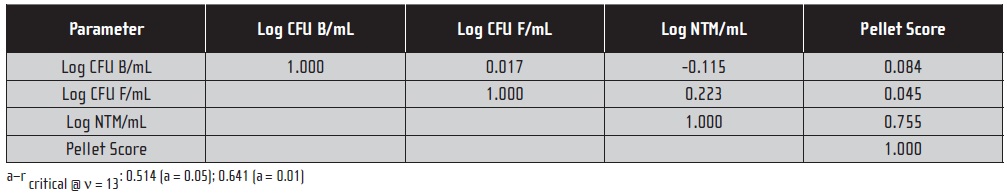
When bacteria and NTM data are compared among samples that had ≥ 2 CFU NTM/mL, the apparent positive correlation disappears (see Table 3). The only significant correlation in this subset is the one between NTM and AFB. These results support the hypothesis that the apparent relationship between bacteria and NTM in the full data set was driven by the substantial number of samples for which all test results were negative.
Table 3. Correlation Matrix for Microbiological Parameters from 15-Sample Metalworking Fluid Data Sub-set; Samples with ≥ 2 CFU NTM . mL.-1(a)
More than a dozen commonly recovered bacterial and fungal taxa, commonly recovered from MWFs, are known to induce HP.
CONCLUSIONS
The data collected during the course of this case study demonstrates that there is no relationship between the mycobacteria and non-mycobacteria in MWF. They also support Passman’s argument that undue focus on mycobacteria as the principal agent causing HP among machinists is imprudent (6). Although they have been recovered from some MWF systems near workers who’ve contracted HP, NTM are common MWF contaminants that are recovered from MWF in plants where no respiratory disease has been reported. More than a dozen commonly recovered bacterial and fungal taxa, commonly recovered from MWF, are known to induce HP. Speculation that investigations that failed to recover NTM in MWF where workers were diagnosed with HP had been insufficient should be reconsidered, as should the focus on NTM eradication. When HP clusters occur, the focus should be on overall bioaerosol control.
Fortunately, there have not been any HP clusters reported in the past several years. Should future outbreaks occur, the authors recommend a multidisciplinary approach to the cause and effect investigation. Microbiological culture tests should be augmented with non-culture analysis such as population profiling using polymerase chain reaction methods. Such non-culture based testing will provide a better understanding of the microbial population in the MWF. Fluid testing should be complemented with both aerosol and surface testing. Surface populations are known to recontaminate recirculating MWF. Worker exposure to the antigens that cause HP is through inhalation. Having now established that there is no relationship between NTM and non-mycobacteria in MWF, it’s time to think more globally about preventing microbially induced respiratory disease in the metalworking environment.
REFERENCES
1. Kreiss, K. and Cox-Ganser, J. (1997), “Metalworking Fluid-Associated Hypersensitivity Pneumonitis: A Workshop Summary,” Am. J. Ind. Med., 32(4), pp. 423-432.
2. Passman, F.J. and Rossmoore, H.W. (2002), “Reassessing the Health Risks Associated with Employee Exposure to Metalworking Fluid Microbes,” Lubrication Engineering, 58(7), pp. 30-38.
3. Watt, W. D. (2003), “Observations on the Relationship Between Triazines and Mycobacteria in Metalworking Fluids,” Appl. Occ. Env. Hyg., 18(11), pp. 961-965.
4. 9215A.6 (2004), “Heterotrophic Plate Count,” In: Standard Methods for Water and Wastewater Analysis, 20th Edition, American Public Health Association, Washington, D.C.
5. E2564-07 (2007), “Standard Test Method for Enumeration of Mycobacteria by Direct Microscopic Counting (DMC) Method,” ASTM International, West Conshohocken, Pa.
6. Passman, F.J. (2008), “Metalworking Fluid Microbes – What We need to Know to Successfully Understand Cause and Effect Relationships,” Tribology Transactions, 51(1), pp. 107-117.
STLE Fellow Fred Passman is president of Biodeterioration Control Associates, Inc., Princeton N.J. You can reach him at fredp@biodeterioration-control.com. STLE-member Katalin Rossmoore is with Biosan Laboratories in Warren, Mich. You can reach her at kati@biosan.com. STLE-member Leonard Rossmoore also works with Biosan Laboratories. You can reach him at len@biosan.com.